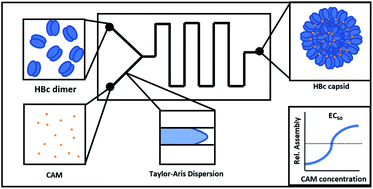
To date, hepatitis B virus (HBV) capsid assembly modulators (CAMs), which target the viral core protein and induce the formation of non-functional viral capsids, have been identified and characterized in microtiter plate-based biochemical or cell-based in vitro assays. In this work, we developed an automated microfluidic screening assay, which uses convection-dominated Taylor–Aris dispersion to generate high-resolution dose–response curves, enabling the measurements of compound EC50 values at very short incubation times. The measurement of early kinetics down to 7.7 seconds in the microfluidic format was utilized to discriminate between the two different classes of CAMs known so far. The CAM (-N), leading to the formation of morphologically normal capsids and the CAM (-A), leading to aberrant HBV capsid structures. CAM-A compounds like BAY 41-4109 and GLS4 showed rapid kinetics, with assembly rates above 80% of the core protein after only a 7 second exposure to the compound, whereas CAM-N compounds like ABI-H0731 and JNJ-56136379 showed significantly slower kinetics. Using our microfluidic system, we characterized two of our in-house screening compounds. Interestingly, one compound showed a CAM-N/A intermediate behavior, which was verified with two standard methods for CAM classification, size exclusion chromatography, and anti-HBc immunofluorescence microscopy. With this proof-of-concept study, we believe that this microfluidic system is a robust primary screening tool for HBV CAM drug discovery, especially for the hit finding and hit-to-lead optimization phases. In addition to EC50 values, this system gives valuable first information about the mode of action of novel CAM screening compounds.